The Normal Flora In a healthy animal, the internal tissues, e.g. blood, brain, muscle, etc., are normally free of microorganisms. However, the surface tissues, i.e., skin and mucous membranes, are constantly in contact with environmental organisms and become readily colonized by various microbial species. The mixture of organisms regularly found at any anatomical site is referred to as the normal flora, except by researchers in the field who prefer the term "indigenous microbiota". The normal flora of humans consists of a few eucaryotic fungi and protists, but bacteria are the most numerous and obvious microbial components of the normal flora.
 Figure 1. Gram stain of a species of Micrococcus, commonly isolated from the skin and nasal membranes of humans. The predominant bacterial flora of humans are shown in Table 1. This table lists only a fraction of the total bacterial species that occur as normal flora of humans. A recent experiment that used 16S RNA probes to survey the diversity of bacteria in dental plaque revealed that only one percent of the total species found have ever been cultivated. Similar observations have been made with the intestinal flora. Also, this table does not indicate the relative number or concentration of bacteria at a particular site. If you are reading online, you can skip this table and use it as an ongoing reference. To continue this article, scroll to the bottom of the Table notes toAssociations Between Humans and the Normal Flora
Figure 1. Gram stain of a species of Micrococcus, commonly isolated from the skin and nasal membranes of humans. The predominant bacterial flora of humans are shown in Table 1. This table lists only a fraction of the total bacterial species that occur as normal flora of humans. A recent experiment that used 16S RNA probes to survey the diversity of bacteria in dental plaque revealed that only one percent of the total species found have ever been cultivated. Similar observations have been made with the intestinal flora. Also, this table does not indicate the relative number or concentration of bacteria at a particular site. If you are reading online, you can skip this table and use it as an ongoing reference. To continue this article, scroll to the bottom of the Table notes toAssociations Between Humans and the Normal Flora
Table 1. Bacteria commonly found on the surfaces of the human body.
| BACTERIUM | Skin | Con-
junc-
tiva | Nose | Pharynx | Mouth | Lower
GI | Ant. ure-
thra | Vagina |
| Staphylococcus epidermidis (1) | ++ | + | ++ | ++ | ++ | + | ++ | ++ |
| Staphylococcus aureus* (2) | + | +/- | + | + | + | ++ | +/- | + |
| Streptococcus mitis |
|
|
| + | ++ | +/- | + | + |
| Streptococcus salivarius |
|
|
| ++ | ++ | |
|
|
| Streptococcus mutans* (3) |
|
|
| + | ++ |
|
|
|
| Enterococcus faecalis* (4) |
|
|
| +/- | + | ++ | + | + |
| Streptococcus pneumoniae* (5) |
| +/- | +/- | + | + |
|
| +/- |
| Streptococcus pyogenes* (6) | +/- | +/- |
| + | + | +/- |
| +/- |
| Neisseria sp. (7) |
| + | + | ++ | + |
| + | + |
| Neisseria meningitidis* (8) |
|
| + | ++ | + |
|
| + |
| Enterobacteriaceae*(Escherichia coli) (9) |
| +/- | +/- | +/- | + | ++ | + | + |
| Proteus sp. |
| +/- | + | + | + | + | + | + |
| Pseudomonas aeruginosa* (10) |
|
|
| +/- | +/- | + | +/- |
|
| Haemophilus influenzae* (11) |
| +/- | + | + | + |
|
|
|
| Bacteroides sp.* |
|
|
|
|
| ++ | + | +/- |
| Bifidobacterium bifidum (12) |
|
|
|
|
| ++ |
|
|
| Lactobacillus sp. (13) |
|
|
| + | ++ | ++ |
| ++ |
| Clostridium sp.* (14) |
|
|
|
| +/- | ++ |
|
|
| Clostridium tetani (15) |
|
|
|
|
| +/- |
|
|
| Corynebacteria (16) | ++ | + | ++ | + | + | + | + | + |
| Mycobacteria | + |
| +/- | +/- |
| + | + |
|
| Actinomycetes |
|
|
| + | + |
|
|
|
| Spirochetes |
|
|
| + | ++ | ++ |
|
|
| Mycoplasmas |
|
|
| + | + | + | +/- | + |
++ = nearly 100 percent + = common (about 25 percent) +/- = rare (less than 5%) * = potential pathogen
Table 1 Notes(1) The staphylococci and corynebacteria occur at every site listed.Staphylococcus epidermidis is highly adapted to the diverse environments of its human host. S. aureus is a potential pathogen. It is a leading cause of bacterial disease in humans. It can be transmitted from the nasal membranes of an asymptomatic carrier to a susceptible host.

S. epidermidis. Scanning EM. CDC.(2) Many of the normal flora are either pathogens or opportunistic pathogens, The asterisks indicate members of the normal flora a that may be considered major pathogens of humans.

S. aureus. Gram stain.
(3) Streptococcus mutans is the primary bacterium involved in plaque formation and initiation of dental caries. Viewed as an opportunistic infection, dental disease is one of the most prevalent and costly infectious diseases in the United States.

Streptococcus mutans. Gram stain. CDC
(4) Enterococcus faecalis was formerly classified as Streptococcus faecalis.The bacterium is such a regular a component of the intestinal flora, that many European countries use it as the standard indicator of fecal pollution, in the same way we use E. coli in the U.S. In recent years, Enterococcus faecalis has emerged as a significant, antibiotic-resistant, nosocomial pathogen.

Vancomycin Resistant Enterococcus faecalis. Scanning E.M. CDC
(5) Streptococcus pneumoniae is present in the upper respiratory tract of about half the population. If it invades the lower respiratory tract it can cause pneumonia. Streptococcus pneumoniae causes 95 percent of all bacterial pneumonia.

Streptococcus pneumoniae. Direct fluorescent antibody stain. CDC.
(6) Streptococcus pyogenes refers to the Group A, Beta-hemolytic streptococci. Streptococci cause tonsillitis (strep throat), pneumonia, endocarditis. Some streptococcal diseases can lead to rheumatic fever or nephritis which can damage the heart and kidney.

Streptococcus pyogenes. Gram stain.
(7) Neisseria and other Gram-negative cocci are frequent inhabitants of the upper respiratory tract, mainly the pharynx. Neisseria meningitidis, an important cause of bacterial meningitis, can colonize as well, until the host can develop active immunity against the pathogen.

Neisseria meningitidis. Gram stain.
(8) While E. coli is a consistent resident of the small intestine, many other enteric bacteria may reside here as well, including Klebsiella, Enterobacter andCitrobacter. Some strains of E. coli are pathogens that cause intestinal infections, urinary tract infections and neonatal meningitis.

E. coli. Scanning E.M. Shirley Owens. Center for Electron Optics. Michigan State University.
(9) Pseudomonas aeruginosa is the quintessential opportunistic pathogen of humans that can invade virtually any tissue. It is a leading cause of hospital-acquired (nosocomial) Gram-negative infections, but its source is often exogenous (from outside the host).

Colonies of Pseudomonas aeruginosa growing on an agar plate. The most virulent Pseudomonas species produce mucoid colonies and green pigments such as this isolate.
(10) Haemophilus influenzae is a frequent secondary invader to viral influenza, and was named accordingly. The bacterium was the leading cause of meningitis in infants and children until the recent development of the Hflu type B vaccine.

Haemophilus influenzae. Gram stain.
(11) The greatest number of bacteria are found in the lower intestinal tract, specifically the colon and the most prevalent bacteria are the Bacteroides, a group of Gram-negative, anaerobic, non-sporeforming bacteria. They have been implicated in the initiation colitis and colon cancer.

Bacteroides fragilis. Gram stain.
(12) Bifidobacteria are Gram-positive, non-sporeforming, lactic acid bacteria. They have been described as "friendly" bacteria in the intestine of humans.Bifidobacterium bifidum is the predominant bacterial species in the intestine of breast-fed infants, where it presumably prevents colonization by potential pathogens. These bacteria are sometimes used in the manufacture of yogurts and are frequently incorporated into probiotics.

Bifidobacterium bifidum. Gram stain
(13) Lactobacilli in the oral cavity probably contribute to acid formation that leads to dental caries. Lactobacillus acidophilus colonizes the vaginal epithelium during child-bearing years and establishes the low pH that inhibits the growth of pathogens.

Lactobacillus species and a vaginal squaemous epithelial cell. CDC
(14) There are numerous species of Clostridium that colonize the bowel.Clostridium perfringens is commonly isolated from feces. Clostridium difficilemay colonize the bowel and cause "antibiotic-induced diarrhea" or pseudomembranous colitis.

Clostridium perfringens. Gram stain.
(15) Clostridium tetani is included in the table as an example of a bacterium that is "transiently associated" with humans as a component of the normal flora. The bacterium can be isolated from feces in 0 - 25 percent of the population. The endospores are probably ingested with food and water, and the bacterium does not colonize the intestine.

Clostridium tetani. Gram stain.
(16) The corynebacteria, and certain related propionic acid bacteria, are consistent skin flora. Some have been implicated as a cause of acne.Corynebacterium diphtheriae, the agent of diphtheria, was considered a member of the normal flora before the widespread use of the diphtheria toxoid, which is used to immunize against the disease.

Corynebacterium diphtheriae. No longer a part of the normal flora.
Associations Between Humans and the Normal Flora E. coli is the best known bacterium that regularly associates itself with humans, being an invariable component of the human intestinal tract. Even though E. coliis the most studied of all bacteria, and we know the exact location and sequence of 4,288 genes on its chromosome, we do not fully understand its ecological relationship with humans. In fact, not much is known about the nature of the associations between humans and their normal flora, but they are thought to be dynamic interactions rather than associations of mutual indifference. Both host and bacteria are thought to derive benefit from each other, and the associations are, for the most part,mutualistic. The normal flora derive from their host a steady supply of nutrients, a stable environment, and protection and transport. The host obtains from the normal flora certain nutritional and digestive benefits, stimulation of the development and activity of immune system, and protection against colonization and infection by pathogenic microbes.While most of the activities of the normal flora benefit their host, some of the normal flora are parasitic (live at the expense of their host), and some arepathogenic (capable of producing disease). Diseases that are produced by the normal flora in their host may be called endogenous diseases. Most endogenous bacterial diseases are opportunistic infections, meaning that the the organism must be given a special opportunity of weakness or let-down in the host defenses in order to infect. An example of an opportunistic infection is chronic bronchitis in smokers wherein normal flora bacteria are able to invade the weakened lung. Sometimes the relationship between a member of the normal flora an its host cannot be deciphered. Such a relationship where there is no apparent benefit or harm to either organism during their association is referred to as a commensal relationship. Many of the normal flora that are not predominant in their habitat, even though always present in low numbers, are thought of as commensal bacteria. However, if a presumed commensal relationship is studied in detail, parasitic or mutualistic characteristics often emerge.
Tissue specificity
Most members of the normal bacterial flora prefer to colonize certain tissues and not others. This "tissue specificity" is usually due to properties of both the host and the bacterium. Usually, specific bacteria colonize specific tissues by one or another of these mechanisms.
1. Tissue tropism is the bacterial preference or predilection for certain tissues for growth. One explanation for tissue tropism is that the host provides essential nutrients and growth factors for the bacterium, in addition to suitable oxygen, pH, and temperature for growth.

Lactobacillus acidophilus, informally known as "Doderlein's bacillus" colonizes the vagina because glycogen is produced which provides the bacteria with a source of sugar that they ferment to lactic acid.
2. Specific adherence Most bacteria can colonize a specific tissue or site because they can adhere to that tissue or site in a specific manner that involves complementary chemical interactions between the two surfaces. Specific adherence involves biochemical interactions between bacterial surface components (ligands or adhesins) and host cell molecular receptors. The bacterial components that provide adhesins are molecular parts of their capsules, fimbriae, or cell walls. The receptors on human cells or tissues are usually glycoprotein molecules located on the host cell or tissue surface.  Figure 2. Specific adherence involves complementary chemical interactions between the host cell or tissue surface and the bacterial surface. In the language of medical microbiologist, a bacterial "adhesin" attaches covalently to a host "receptor" so that the bacterium "docks" itself on the host surface. The adhesins of bacterial cells are chemical components of capsules, cell walls, pili or fimbriae. The host receptors are usually glycoproteins located on the cell membrane or tissue surface. Some examples of adhesins and attachment sites used for specific adherence to human tissues are described in the table below.
Figure 2. Specific adherence involves complementary chemical interactions between the host cell or tissue surface and the bacterial surface. In the language of medical microbiologist, a bacterial "adhesin" attaches covalently to a host "receptor" so that the bacterium "docks" itself on the host surface. The adhesins of bacterial cells are chemical components of capsules, cell walls, pili or fimbriae. The host receptors are usually glycoproteins located on the cell membrane or tissue surface. Some examples of adhesins and attachment sites used for specific adherence to human tissues are described in the table below.
Table 2. Examples of bacterial specific adherence to host cells or tissue.
| Bacterium | Bacterial adhesin | Attachment site |
| Streptococcus pyogenes | Cell-bound protein (M-protein) | Pharyngeal epithelium |
| Streptococcus mutans | Cell- bound protein (Glycosyl transferase) | Pellicle of tooth |
| Streptococcus salivarius | Lipoteichoic acid | Buccal epithelium of tongue |
| Streptococcus pneumoniae | Cell-bound protein (choline-binding protein) | Mucosal epithelium |
| Staphylococcus aureus | Cell-bound protein | Mucosal epithelium |
| Neisseria gonorrhoeae | N-methylphenyl- alanine pili | Urethral/cervical epithelium |
| Enterotoxigenic E. coli | Type-1 fimbriae | Intestinal epithelium |
| Uropathogenic E. coli | P-pili (pap) | Upper urinary tract |
| Bordetella pertussis | Fimbriae ("filamentous hemagglutinin") | Respiratory epithelium |
| Vibrio cholerae | N-methylphenylalanine pili | Intestinal epithelium |
| Treponema pallidum | Peptide in outer membrane | Mucosal epithelium |
| Mycoplasma | Membrane protein | Respiratory epithelium |
| Chlamydia | Unknown | Conjunctival or urethral epithelium |
|
3. Biofilm formation Some of the indigenous bacteria are able to construct biofilms on a tissue surface, or they are able to colonize a biofilm built by another bacterial species. Many biofilms are a mixture of microbes, although one member is responsible for maintaining the biofilm and may predominate. Figure 3. Cartoon depicting biofilm formation. Biofilms usually occur when one bacterial species attaches specifically or non specifically to a surface, and then secretes carbohydrate slime (exopolymer) that imbeds the bacteria and attracts other microbes to the biofilm for protection or nutritional advantages. The classic biofilm that involves components of the normal flora of the oral cavity is the formation of dental plaque on the teeth. Plaque is a naturally-constructed biofilm, in which the consortia of bacteria may reach a thickness of 300-500 cells on the surfaces of the teeth. These accumulations subject the teeth and gingival tissues to high concentrations of bacterial metabolites, which result in dental disease.
The Composition of the Normal Flora The normal flora of humans are exceedingly complex and consist of more than 200 species of bacteria. The makeup of the normal flora may be influenced by various factors, including genetics, age, sex, stress, nutrition and diet of the individual. Three developmental changes in humans, weaning, the eruption of the teeth, and the onset and cessation of ovarian functions, invariably affect the composition of the normal flora in the intestinal tract, the oral cavity, and the vagina, respectively. However, within the limits of these fluctuations, the bacterial flora of humans is sufficiently constant to a give general description of the situation.
Figure 3. Cartoon depicting biofilm formation. Biofilms usually occur when one bacterial species attaches specifically or non specifically to a surface, and then secretes carbohydrate slime (exopolymer) that imbeds the bacteria and attracts other microbes to the biofilm for protection or nutritional advantages. The classic biofilm that involves components of the normal flora of the oral cavity is the formation of dental plaque on the teeth. Plaque is a naturally-constructed biofilm, in which the consortia of bacteria may reach a thickness of 300-500 cells on the surfaces of the teeth. These accumulations subject the teeth and gingival tissues to high concentrations of bacterial metabolites, which result in dental disease.
The Composition of the Normal Flora The normal flora of humans are exceedingly complex and consist of more than 200 species of bacteria. The makeup of the normal flora may be influenced by various factors, including genetics, age, sex, stress, nutrition and diet of the individual. Three developmental changes in humans, weaning, the eruption of the teeth, and the onset and cessation of ovarian functions, invariably affect the composition of the normal flora in the intestinal tract, the oral cavity, and the vagina, respectively. However, within the limits of these fluctuations, the bacterial flora of humans is sufficiently constant to a give general description of the situation.
A human first becomes colonized by a normal flora at the moment of birth and passage through the birth canal. In utero, the fetus is sterile, but when the mother's water breaks and the birth process begins, so does colonization of the body surfaces. Handling and feeding of the infant after birth leads to establishment of a stable normal flora on the skin, oral cavity and intestinal tract in about 48 hours.
It has been calculated that a human adult houses about 1012 bacteria on the skin, 1010 in the mouth, and 1014 in the gastrointestinal tract. The latter number is far in excess of the number of eucaryotic cells in all the tissues and organs which comprise a human. The predominant bacteria on the surfaces of the human body are listed in Table 3. Informal names identify the bacteria in this table. Formal taxonomic names of organisms are given in Table 1.
Table 3. Predominant bacteria at various anatomical locations in adults.
| Anatomical Location | Predominant bacteria |
| Skin | staphylococci and corynebacteria |
| Conjunctiva | sparse, Gram-positive cocci and Gram-negative rods |
| Oral cavity |
|
| teeth | streptococci, lactobacilli |
| mucous membranes | streptococci and lactic acid bacteria |
| Upper respiratory tract |
|
| nares (nasal membranes) | staphylococci and corynebacteria |
| pharynx (throat) | streptococci, neisseria, Gram-negative rods and cocci |
| Lower respiratory tract | none |
| Gastrointestinal tract |
|
| stomach | Helicobacter pylori (up to 50%) |
| small intestine | lactics, enterics, enterococci, bifidobacteria |
| colon | bacteroides, lactics, enterics, enterococci, clostridia, methanogens |
| Urogenital tract |
|
| anterior urethra | sparse, staphylococci, corynebacteria, enterics |
| vagina | lactic acid bacteria during child-bearing years; otherwise mixed |
Normal Flora of the Skin The adult human is covered with approximately 2 square meters of skin. The density and composition of the normal flora of the skin varies with anatomical locale. The high moisture content of the axilla, groin, and areas between the toes supports the activity and growth of relatively high densities of bacterial cells, but the density of bacterial populations at most other sites is fairly low, generally in 100s or 1000s per square cm. Most bacteria on the skin are sequestered in sweat glands.
The skin microbes found in the most superficial layers of the epidermis and the upper parts of the hair follicles are Gram-positive cocci (Staphylococcus epidermidis and Micrococcus sp.) and corynebacteria such as Propionibacteriumsp. These are generally nonpathogenic and considered to be commensal, although mutualistic and parasitic roles have been assigned to them. For example, staphylococci and propionibacteria produce fatty acids that inhibit the growth of fungi and yeast on the skin. But, if Propionibacterium acnes, a normal inhabitant of the skin, becomes trapped in hair follicle, it may grow rapidly and cause inflammation and acne.
Sometimes potentially pathogenic Staphylococcus aureus is found on the face and hands in individuals who are nasal carriers. This is because the face and hands are likely to become inoculated with the bacteria on the nasal membranes. Such individuals may autoinoculate themselves with the pathogen or spread it to other individuals or foods.Normal Flora of the Conjunctiva A variety of bacteria may be cultivated from the normal conjunctiva, but the number of organisms is usually small.Staphylococcus epidermidis and certain coryneforms (Propionibacterium acnes) are dominant. Staphylococcus aureus, some streptococci, Haemophilus sp. andNeisseria sp. are occasionally found. The conjunctiva is kept moist and healthy by the continuous secretions from the lachrymal glands. Blinking wipes the conjunctiva every few seconds mechanically washing away foreign objects including bacteria. Lachrymal secretions (tears) also contain bactericidal substances including lysozyme. There is little or no opportunity for microorganisms to colonize the conjunctiva without special mechanisms to attach to the epithelial surfaces and some ability to withstand attack by lysozyme.Pathogens which do infect the conjunctiva (e.g. Neisseria gonorrhoeae andChlamydia trachomatis) are thought to be able to specifically attach to the conjunctival epithelium. Newborn infants may be especially prone to bacterial attachment. Since Chlamydia and Neisseria might be present on the cervical and vaginal epithelium of an infected mother, silver nitrate or an antibiotic may be put into the newborn's eyes to avoid infection after passage through the birth canal.
 Figure 4. Colonies of Propionibacterium acnes, found on skin and the conjunctiva.
Figure 4. Colonies of Propionibacterium acnes, found on skin and the conjunctiva.
Normal Flora of the Respiratory Tract A large number of bacterial species colonize the upper respiratory tract (nasopharynx). The nares (nostrils) are always heavily colonized, predominantly with Staphylococcus epidermidis and corynebacteria, and often (in about 20% of the general population) withStaphylococcus aureus, this being the main carrier site of this important pathogen. The healthy sinuses, in contrast are sterile. The pharynx (throat) is normally colonized by streptococci and various Gram-negative cocci. Sometimes pathogens such as Streptococcus pneumoniae, Streptococcus pyogenes, Haemophilus influenzae and Neisseria meningitidis colonize the pharynx.
The lower respiratory tract (trachea, bronchi, and pulmonary tissues) is virtually free of microorganisms, mainly because of the efficient cleansing action of the ciliated epithelium which lines the tract. Any bacteria reaching the lower respiratory tract are swept upward by the action of the mucociliary blanket that lines the bronchi, to be removed subsequently by coughing, sneezing, swallowing, etc. If the respiratory tract epithelium becomes damaged, as in bronchitis or viral pneumonia, the individual may become susceptible to infection by pathogens such as H. influenzae or S. pneumoniae descending from the nasopharynx.
Normal Flora of the Urogenital Tract Urine is normally sterile, and since the urinary tract is flushed with urine every few hours, microorganisms have problems gaining access and becoming established. The flora of the anterior urethra, as indicated principally by urine cultures, suggests that the area my be inhabited by a relatively consistent normal flora consisting of Staphylococcus epidermidis, Enterococcus faecalis and some alpha-hemolytic streptococci. Their numbers are not plentiful, however. In addition, some enteric bacteria (e.g. E. coli, Proteus) and corynebacteria, which are probably contaminants from the skin, vulva or rectum, may occasionally be found at the anterior urethra.
The vagina becomes colonized soon after birth with corynebacteria, staphylococci, streptococci, E. coli, and a lactic acid bacterium historically named "Doderlein's bacillus" (Lactobacillus acidophilus). During reproductive life, from puberty to menopause, the vaginal epithelium contains glycogen due to the actions of circulating estrogens. Doderlein's bacillus predominates, being able to metabolize the glycogen to lactic acid. The lactic acid and other products of metabolism inhibit colonization by all except this lactobacillus and a select number of lactic acid bacteria. The resulting low pH of the vaginal epithelium prevents establishment by most other bacteria as well as the potentially-pathogenic yeast, Candida albicans. This is a striking example of the protective effect of the normal bacterial flora for their human host.
 Figure 5. A Lactobacillus species, possibly Doderlein's bacillus, in association with a vaginal epithelial cell.
Figure 5. A Lactobacillus species, possibly Doderlein's bacillus, in association with a vaginal epithelial cell.
Normal Flora of the Oral Cavity The presence of nutrients, epithelial debris, and secretions makes the mouth a favorable habitat for a great variety of bacteria. Oral bacteria include streptococci, lactobacilli, staphylococci and corynebacteria, with a great number of anaerobes, especially bacteroides.
The mouth presents a succession of different ecological situations with age, and this corresponds with changes in the composition of the normal flora. At birth, the oral cavity is composed solely of the soft tissues of the lips, cheeks, tongue and palate, which are kept moist by the secretions of the salivary glands. At birth the oral cavity is sterile but rapidly becomes colonized from the environment, particularly from the mother in the first feeding. Streptococcus salivarius is dominant and may make up 98% of the total oral flora until the appearance of the teeth (6 - 9 months in humans). The eruption of the teeth during the first year leads to colonization by S. mutans and S. sanguis. These bacteria require a nondesquamating (nonepithelial) surface in order to colonize. They will persist as long as teeth remain. Other strains of streptococci adhere strongly to the gums and cheeks but not to the teeth. The creation of the gingival crevice area (supporting structures of the teeth) increases the habitat for the variety of anaerobic species found. The complexity of the oral flora continues to increase with time, and bacteroides and spirochetes colonize around puberty.
 Figure 6. Various streptococci in a biofilm in the oral cavity.
Figure 6. Various streptococci in a biofilm in the oral cavity.
The normal bacterial flora of the oral cavity clearly benefit from their host who provides nutrients and habitat. There may be benefits, as well, to the host. The normal flora occupy available colonization sites which makes it more difficult for other microorganisms (nonindigenous species) to become established. Also, the oral flora contribute to host nutrition through the synthesis of vitamins, and they contribute to immunity by inducing low levels of circulating and secretory antibodies that may cross react with pathogens. Finally, the oral bacteria exert microbial antagonism against nonindigenous species by production of inhibitory substances such as fatty acids, peroxides and bacteriocins.
On the other hand, the oral flora are the usual cause of various oral diseases in humans, including abscesses, dental caries, gingivitis, and periodontal disease. If oral bacteria can gain entrance into deeper tissues, they may cause abscesses of alveolar bone, lung, brain, or the extremities. Such infections usually contain mixtures of bacteria with Bacteroides melaninogenicus often playing a dominant role. If oral streptococci are introduced into wounds created by dental manipulation or treatment, they may adhere to heart valves and initiate subacute bacterial endocarditis.
 Figure 7. Colonies of E. coli growing on EMB agar
Figure 7. Colonies of E. coli growing on EMB agar.
Normal Flora of the Gastrointestinal Tract The bacterial flora of the gastrointestinal (GI) tract of animals has been studied more extensively than that of any other site. The composition differs between various animal species, and within an animal species. In humans, there are differences in the composition of the flora which are influenced by age, diet, cultural conditions, and the use of antibiotics. The latter greatly perturbs the composition of the intestinal flora.
In the upper GI tract of adult humans, the esophagus contains only the bacteria swallowed with saliva and food. Because of the high acidity of the gastric juice, very few bacteria (mainly acid-tolerant lactobacilli) can be cultured from the normal stomach. However, at least half the population in the United States is colonized by a pathogenic bacterium, Helicobacter pylori. Since the 1980s, this bacterium has been known to be the cause of gastric ulcers, and it is probably a cause of gastric and duodenal cancer as well. The Australian microbiologist, Barry Marshall, received the Nobel Prize in Physiology and Medicine in 2005, for demonstrating the relationship between Helicobacter and gastric ulcers.
 Figure 8. Helicobacter pylori. ASM
Figure 8. Helicobacter pylori. ASM
The proximal small intestine has a relatively sparse Gram-positive flora, consisting mainly of lactobacilli and Enterococcus faecalis. This region has about 105 - 107 bacteria per ml of fluid. The distal part of the small intestine contains greater numbers of bacteria (108/ml) and additional species, including coliforms (E. coli and relatives) and Bacteroides, in addition to lactobacilli and enterococci.
The flora of the large intestine (colon) is qualitatively similar to that found in feces. Populations of bacteria in the colon reach levels of 1011/ml feces. Coliforms become more prominent, and enterococci, clostridia and lactobacilli can be regularly found, but the predominant species are anaerobic Bacteroidesand anaerobic lactic acid bacteria in the genus Bifidobacterium (Bifidobacterium bifidum). These organisms may outnumber E. coli by 1,000:1 to 10,000:1. Sometimes, significant numbers of anaerobic methanogens (up to 1010/gm) may reside in the colon of humans. This is our only direct association with archaea as normal flora. The range of incidence of certain bacteria in the large intestine of humans is shown in Table 4 below.
Table 4. Bacteria found in the large intestine of humans.
| BACTERIUM | RANGE OF INCIDENCE |
| Bacteroides fragilis | 100 |
| Bacteroides melaninogenicus | 100 |
| Bacteroides oralis | 100 |
| Lactobacillus | 20-60 |
| Clostridium perfringens | 25-35 |
| Clostridium septicum | 5-25 |
| Clostridium tetani | 1-35 |
| Bifidobacterium bifidum | 30-70 |
| Staphylococcus aureus | 30-50 |
| Enterococcus faecalis | 100 |
| Escherichia coli | 100 |
| Salmonella enteritidis | 3-7 |
| Klebsiella sp. | 40-80 |
| Enterobacter sp. | 40-80 |
| Proteus mirabilis | 5-55 |
| Pseudomonas aeruginosa | 3-11 |
| Peptostreptococcus sp. | ?common |
| Peptococcus sp. | ?common |
At birth the entire intestinal tract is sterile, but bacteria enter with the first feed. The initial colonizing bacteria vary with the food source of the infant. In breast-fed infants, bifidobacteria account for more than 90% of the total intestinal bacteria. Enterobacteriaceae and enterococci are regularly present, but in low proportions, while bacteroides, staphylococci, lactobacilli and clostridia are practically absent. In bottle-fed infants, bifidobacteria are not predominant. When breast-fed infants are switched to a diet of cow's milk or solid food, bifidobacteria are progressively joined by enterics, bacteroides, enterococci lactobacilli and clostridia. Apparently, human milk contains a growth factor that enriches for growth of bifidobacteria, and these bacteria play an important role in preventing colonization of the infant intestinal tract by non indigenous or pathogenic species. 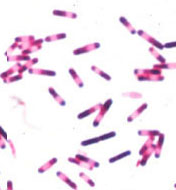 Figure 9. Clostridium difficile. Gram stain. The growth of "C. diff" in the intestinal tract is normally held in check by other members of the normal flora. When antibiotics given for other infections cause collateral damage to the normal intestinal flora, the clostridium may be able to "grow out" and produce a serious diarrheal syndrome called pseudomembranous colitis. This is an example of an "antibiotic induced diarrheal disease".The composition of the flora of the gastrointestinal tract varies along the tract (at longitudinal levels) and across the tract (at horizontal levels) where certain bacteria attach to the gastrointestinal epithelium and others occur in the lumen. There is frequently a very close association between specific bacteria in the intestinal ecosystem and specific gut tissues or cells (evidence of tissue tropism and specific adherence). Gram-positive bacteria, such as the streptococci and lactobacilli, are thought to adhere to the gastrointestinal epithelium using polysaccharide capsules or cell wall teichoic acids to attach to specific receptors on the epithelial cells. Gram-negative bacteria such as the enterics may attach by means of specific fimbriae which bind to glycoproteins on the epithelial cell surface.
Figure 9. Clostridium difficile. Gram stain. The growth of "C. diff" in the intestinal tract is normally held in check by other members of the normal flora. When antibiotics given for other infections cause collateral damage to the normal intestinal flora, the clostridium may be able to "grow out" and produce a serious diarrheal syndrome called pseudomembranous colitis. This is an example of an "antibiotic induced diarrheal disease".The composition of the flora of the gastrointestinal tract varies along the tract (at longitudinal levels) and across the tract (at horizontal levels) where certain bacteria attach to the gastrointestinal epithelium and others occur in the lumen. There is frequently a very close association between specific bacteria in the intestinal ecosystem and specific gut tissues or cells (evidence of tissue tropism and specific adherence). Gram-positive bacteria, such as the streptococci and lactobacilli, are thought to adhere to the gastrointestinal epithelium using polysaccharide capsules or cell wall teichoic acids to attach to specific receptors on the epithelial cells. Gram-negative bacteria such as the enterics may attach by means of specific fimbriae which bind to glycoproteins on the epithelial cell surface.
It is in the intestinal tract that we see the greatest effect of the bacterial flora on their host. This is due to their large mass and numbers. Bacteria in the human GI tract have been shown to produce vitamins and may otherwise contribute to nutrition and digestion. But their most important effects are in their ability to protect their host from establishment and infection by alien microbes and their ability to stimulate the development and the activity of the immunological tissues.
On the other hand, some of the bacteria in the colon (e.g. Bacteroides) have been shown to produce metabolites that are carcinogenic, and there may be an increased incidence of colon cancer associated with these bacteria. Alterations in the GI flora brought on by poor nutrition or perturbance with antibiotics can cause shifts in populations and colonization by nonresidents that leads to gastrointestinal disease.
Beneficial Effects of the Normal Flora
The effects of the normal flora are inferred by microbiologists from experimental comparisons between "germ-free" animals (which are not colonized by any microbes) and conventional animals (which are colonized with a typical normal flora). Briefly, some of the characteristics of a germ-free animals that are thought to be due to lack of exposure to a normal flora are:
1. vitamin deficiencies, especially vitamin K and vitamin B12
2. increased susceptibility to infectious disease
3. poorly developed immune system, especially in the gastrointestinal tract
4. lack of "natural antibody" or natural immunity to bacterial infection
Because these conditions in germ-free mice and hamsters do not occur in conventional animals, or are alleviated by introduction of a bacterial flora (at the appropriate time of development), it is tempting to conclude that the human normal flora make similar contributions to human nutrition, health and development. The overall beneficial effects of microbes are summarized below.
1. The normal flora synthesize and excrete vitamins in excess of their own needs, which can be absorbed as nutrients by their host. For example, in humans, enteric bacteria secrete Vitamin K and Vitamin B12, and lactic acid bacteria produce certain B-vitamins. Germ-free animals may be deficient in Vitamin K to the extent that it is necessary to supplement their diets.
2. The normal flora prevent colonization by pathogens by competing for attachment sites or for essential nutrients. This is thought to be their most important beneficial effect, which has been demonstrated in the oral cavity, the intestine, the skin, and the vaginal epithelium. In some experiments, germ-free animals can be infected by 10 Salmonella bacteria, while the infectious dose for conventional animals is near 106 cells.
3. The normal flora may antagonize other bacteria through the production of substances which inhibit or kill nonindigenous species. The intestinal bacteria produce a variety of substances ranging from relatively nonspecific fatty acids and peroxides to highly specific bacteriocins, which inhibit or kill other bacteria.
4. The normal flora stimulate the development of certain tissues, i.e., the caecum and certain lymphatic tissues (Peyer's patches) in the GI tract. The caecum of germ-free animals is enlarged, thin-walled, and fluid-filled, compared to that organ in conventional animals. Also, based on the ability to undergo immunological stimulation, the intestinal lymphatic tissues of germ-free animals are poorly-developed compared to conventional animals.
5. The normal flora stimulate the production of natural antibodies.Since the normal flora behave as antigens in an animal, they induce an immunological response, in particular, an antibody-mediated immune (AMI) response. Low levels of antibodies produced against components of the normal flora are known to cross react with certain related pathogens, and thereby prevent infection or invasion. Antibodies produced against antigenic components of the normal flora are sometimes referred to as "natural" antibodies, and such antibodies are lacking in germ-free animals.
Harmful Effects of the Normal Flora
Harmful effects of the normal flora, some of which are observed in studies with germ-free animals, can be put in the following categories. All but the last two are fairly insignificant.1. Bacterial synergism between a member of the normal flora and a potential pathogen. This means that one organism is helping another to grow or survive. There are examples of a member of the normal flora supplying a vitamin or some other growth factor that a pathogen needs in order to grow. This is calledcross-feeding between microbes. Another example of synergism occurs during treatment of "staph-protected infections" when a penicillin-resistant staphylococcus that is a component of the normal flora shares its drug resistance with pathogens that are otherwise susceptible to the drug.2. Competition for nutrients Bacteria in the gastrointestinal tract must absorb some of the host's nutrients for their own needs. However, in general, they transform them into other metabolisable compounds, but some nutrient(s) may be lost to the host. Germ-free animals are known to grow more rapidly and efficiently than conventional animals. One explanation for incorporating antibiotics into the food of swine, cows and poultry is that the animal grows faster and can therefore be marketed earlier. Unfortunately, this practice contributes to the development and spread of bacterial antibiotic resistance within the farm animals, as well as humans. 3. Induction of a low grade toxemia Minute amounts of bacterial toxins (e.g. endotoxin) may be found in the circulation. Of course, it is these small amounts of bacterial antigen that stimulate the formation of natural antibodies.4. The normal flora may be agents of disease. Members of the normal flora may cause endogenous disease if they reach a site or tissue where they cannot be restricted or tolerated by the host defenses. Many of the normal flora are potential pathogens, and if they gain access to a compromised tissue from which they can invade, disease may result. 5. Transfer to susceptible hosts Some pathogens of humans that are members of the normal flora may also rely on their host for transfer to other individuals where they can produce disease. This includes the pathogens that colonize the upper respiratory tract such as Neisseria meningitidis, Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus, and potential pathogens such as E. coli, Salmonella or Clostridium in the gastrointestinal tract.
Dental Caries, Gingivitis and Periodontal Disease The most frequent and economically-important condition in humans resulting from interactions with our normal flora is probably dental caries. Dental plaque, dental caries, gingivitis and periodontal disease result from actions initiated and carried out by the normal bacterial flora.
Dental plaque, which is material adhering to the teeth, consists of bacterial cells (60-70% the volume of the plaque), salivary polymers, and bacterial extracellular products. Plaque is a naturally-constructed biofilm, in which the consortia of bacteria may reach a thickness of 300-500 cells on the surfaces of the teeth. These accumulations subject the teeth and gingival tissues to high concentrations of bacterial metabolites, which result in dental disease.
The dominant bacterial species in dental plaque are Streptococcus sanguis andStreptococcus mutans, both of which are considered responsible for plaque.
 Streptococcus mutans. Gram stain. CDC
Streptococcus mutans. Gram stain. CDC.
Plaque formation is initiated by a weak attachment of the streptococcal cells to salivary glycoproteins forming a pellicle on the surface of the teeth. This is followed by a stronger attachment by means of extracellular sticky polymers of glucose (glucans) which are synthesized by the bacteria from dietary sugars (principally sucrose). An enzyme on the cell surface of Streptococcus mutans, glycosyl transferase, is involved in initial attachment of the bacterial cells to the tooth surface and in the conversion of sucrose to dextran polymers (glucans) which form plaque.
 Dental plaque, scanning electron micrograph illustrating the diversity of microbes in plaque. Image courtesy of Rachel Sammons, University of Birmingham School of Dentistry (UK).
Dental plaque, scanning electron micrograph illustrating the diversity of microbes in plaque. Image courtesy of Rachel Sammons, University of Birmingham School of Dentistry (UK).
Dental Caries is the destruction of the enamel, dentin or cementum of teeth due to bacterial activities. Caries are initiated by direct demineralization of the enamel of teeth due to lactic acid and other organic acids which accumulate in dental plaque. Lactic acid bacteria in the plaque produce lactic acid from the fermentation of sugars and other carbohydrates in the diet of the host.Streptococcus mutans and Streptococcus sanguis are most consistently been associated with the initiation of dental caries, but other lactic acid bacteria are probably involved as well. These organisms normally colonize the occlusal fissures and contact points between the teeth, and this correlates with the incidence of decay on these surfaces.
Cross section of a tooth illustrating the various structural regions susceptible to colonization or attack by microbes.
Streptococcus mutans in particular has a number of physiological and biochemical properties which implicate it in the initiation of dental caries.
1. It is a regular component of the normal oral flora of humans which occurs in relatively large numbers. It readily colonizes tooth surfaces: salivary components (mucins, which are glycoproteins) form a thin film on the tooth called the enamel pellicle. The adsorbed mucins are thought to serve as molecular receptors for ligands on the bacterial cell surface.2. It contains a cell-bound protein, glycosyl transferase, that serves an adhesin for attachment to the tooth, and as an enzyme that polymerizes dietary sugars into glucans that leads to the formation of plaque.
3. It produces lactic acid from the utilization of dietary carbohydrate which demineralizes tooth enamel. S. mutans produces more lactic acid and is more acid-tolerant than most other streptococci.
4. It stores polysaccharides made from dietary sugars which can be utilized as reserve carbon and energy sources for production of lactic acid. The extracellular glucans formed by S. mutans are, in fact, bacterial capsular polysaccharides that function as carbohydrate reserves. The organisms can also form intracellular polysaccharides from sugars which are stored in cells and then metabolized to lactic acid.
Streptococcus mutans appears to be important in the initiation of dental caries because its activities lead to colonization of the tooth surfaces, plaque formation, and localized demineralization of tooth enamel. It is not however, the only cause of dental decay. After initial weakening of the enamel, various oral bacteria gain access to interior regions of the tooth. Lactobacilli, Actinomyces, and various proteolytic bacteria are commonly found in human carious dentin and cementum, which suggests that they are secondary invaders that contribute to the progression of the lesions.
 Actinomyces israelii
Actinomyces israelii
Periodontal Diseases are bacterial infections that affect the supporting structures of the teeth (gingiva, cementum, periodontal membrane and alveolar bone). The most common form, gingivitis, is an inflammatory condition of the gums. It is associated with accumulations of bacterial plaque in the area. Increased populations of Actinomyces have been found, and they have been suggested as the cause.
Diseases that are confined to the gum usually do not lead to loss of teeth, but there are other more serious forms of periodontal disease that affect periodontal membrane and alveolar bone resulting in tooth loss. Bacteria in these lesions are very complex populations consisting of Gram-positive organisms (includingActinomyces and streptococci) and Gram-negative organisms (including spirochetes and Bacteroides). The mechanisms of tissue destruction in periodontal disease are not clearly defined but hydrolytic enzymes, endotoxins, and other toxic bacterial metabolites seem to be involved.




































